Study design and participants
This exploratory study investigated autoantibodies in patients who had experienced coagulation-related AEFIs secondary to a COVID-19 vaccine, in comparison with patients with other AEFIs, healthy blood donors (BDs), and AEFI cases with COVID-19 exposure. Patients with AEFIs attributed to COVID-19 vaccines were routinely recruited into the SWEDEGENE study (www.swedegene.se). Recruitment followed the standardized SWEDEGENE methodology27. Briefly, we contacted and recruited patients reported to the Swedish Medical Products Agency (MPA) due to a suspected AEFI attributed to a COVID-19 vaccine used in Sweden, which included Comirnaty (Tozinameran, BNT162b2; Pfizer), Spikevax (Elasomeran, mRNA-1273; Moderna), and Vaxzevria (Chimpanzee adenovirus Y25, Covishield, ChAdOx1 nCoV-19, AZD1222; AstraZeneca). Based on available literature reporting vaccination data until October 2021, 82.5% of the population in Sweden had been vaccinated with at least one dose, and 75.7–79% of vaccinated individuals had received at least one dose of Comirnaty, while corresponding percentages for Moderna and Vaxzevria were 9.1–14.1% and 5–15.2%, respectively28,29,30. Causality assessment for AEFIs were performed according to World Health Organization (WHO) criteria, as described previously31. The first subset of samples analyzed in this study were cases with AEFIs that had occurred between December 2020 and September 2021 and had been reported to the MPA between January 2021 and September 2021. The second subset included cases with AEFIs that had occurred until June 2022 that had been reported to the MPA until October 2022 (including available samples from the first subset). An important change in vaccination schedule during the study period was the discontinuation of Vaxzevria use in Sweden after September 2021. As such, the Vaxzevria-attributed AEFI group in the second subset is comprised of subjects from the first subset and also those who received this vaccine prior to its discontinuation but the MPA was notified of the AEFI after September 2021.
The study was approved by the Swedish Ethics Review Board (ethical permit: #2021-06262-01) and all recruitment/analytical processes conformed to the Declaration of Helsinki. All patients provided informed consent and were at least 18 years of age at the time of recruitment. Clinical data (demographics, medical history, drug treatment history, laboratory data, and ancestry) were recorded. Detailed clinical and event-related information concerning individuals with autoantibody positivity for any antigen were collected by re-examining hospitalization records and discharge reports. Time intervals from vaccination to event (AEFI) and event to sampling were recorded.
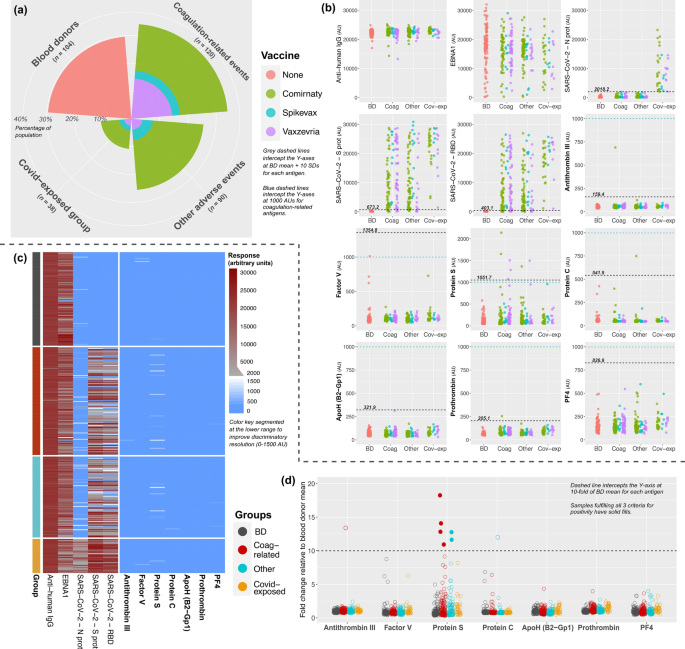
Patients with thrombosis, bleeding, thrombocytopenia or myocardial/cerebral infarction were defined as the ‘coagulation-related AEFI’ group. The healthy control group comprised anonymous BDs who had been sampled before the COVID-19 pandemic at Akademiska Sjukhuset, Uppsala University. The ‘other AEFI’ group included patients with anaphylactic reactions, neurological disorders or peri/myocarditis (Supplementary Table 1). To curtail the potential bias of exposure to COVID-19 itself, we extracted a fourth group comprising COVID-19-exposed subjects. This latter group was created based on elevated antibody response to the nucleocapsid antigen (N protein) of SARS-CoV-2, since the vaccines administered to patients would not create reactivity to this antigen. Any subject who had a nucleocapsid response exceeding the mean value of the BD group by 10 standard deviations (SDs) was included in the ‘covid-exposed’ group regardless of AEFI type.
Analysis subsets
The laboratory analyses were performed in two separate steps since patient recruitment was ongoing throughout the study. The first subset comprised 352 patients, among which 104 were BDs and 248 were from the AEFI cohort (120 coagulation-related AEFI, 90 other AEFI, 38 covid-exposed). These samples underwent antibody screening via a multiplex bead-based assay and confirmation was done by ELISA. The second subset was created by including both newly-received samples (until October 2022) and unthawed samples from the first subset—available for 133 of the subjects (93 of which had coagulation-related AEFI). Ultimately, the second subset comprised 272 individuals, among which 43 were BDs and 229 had AEFIs (186 coagulation-related AEFI, 28 other AEFI, 15 covid-exposed). These samples underwent antibody measurement via in-house and commercial ELISA assays to detect autoantibodies against PF4, PF4-polyanion complexes (PF4C) and antiphospholipid antibodies (ApoH, cardiolipin, and screening). The ‘screening’ refers to the use of the Human Phospholipid Screen IgG/IgM ELISA kit, detailed later on in the text.
Definitions for autoantibody positivity
For the first subset of patients, autoantibody response with the bead-based assay was deemed positive if a particular sample fulfilled three strict criteria: (1) absolute response exceeded the BD mean for that particular antigen by at least 10 SDs, (2) relative response (sample-to-BD mean ratio) exceeded a 10-fold threshold for each antigen, and (3) absolute response was at least 1000 arbitrary units (AUs). Samples fulfilling these criteria were reanalyzed via ELISA to demonstrate elevated response relative to randomly-selected BDs and samples from patients with myocarditis. For the second subset, antibody responses were defined to be positive if they exceeded thresholds created by simultaneously-measured biological samples with known autoantibody positivity for said antigen (or exceeding thresholds defined by commercial kits). When necessary, functional analyses were also performed to confirm the physiological impact of autoantibodies.
Sampling and autoantibody detection
Sampling process
Blood samples were drawn at the patient’s nearest health-care facility (heparinized samples, centrifuged at 1500×g, 10 min, 4 °C), and the resultant plasma was aliquoted and transferred to Uppsala University (stored at −70 °C).
Bead-based immunoassay
A multiplex bead-based immunoassay was performed to detect autoantibodies against multiple target antigens, including the following coagulation-related proteins: Factor V, Protein S, Protein C, Prothrombin, PF4, ApoH (B2-Gp1), and Antithrombin III. Sample detection was confirmed by anti-human IgG response. Antibody response against the Epstein-Barr virus nuclear antigen 1 (EBNA1) was also determined to confirm antibody detection and demonstrate the detection of variabilities in reactivity. Additionally, vaccine response and SARS-CoV-2 exposure were examined by measurement of responses against the Spike (S protein), receptor binding domain (RBD), and nucleocapsid (N protein) antigens.
The first step in the established protocol was the creation of beads coupled to targeted antigens, as detailed previously32,33,34. Magnetic beads (MagPlex®, Luminex) were coupled with commercial, full-length target proteins by use of an AnteoTech activation kit (A-LMPAKMM-10). For each antigen, the protein-to-bead concentration was 3 µg/1.5 × 106 beads. Samples (1 µl) were then diluted 1:250 through a 2-step process: 1:25 in phosphate-buffered saline (PBS) and then 1:10 in PBS containing 0.05% Tween-20, 3% bovine serum albumin (BSA) and 5% non-fat milk. The resultant working samples (250 µl total volume) were incubated with 5 µl of the bead suspension (2 h at room temperature) under slight agitation (orbital shaker at 650 revolutions per minute–RPM). After magnetization and 3 wash cycles (0.05% Tween-20 in PBS), resuspension was performed in 50 µl of 0.2% paraformaldehyde for 10 min, followed by another 3-cycle wash. Secondary antibodies were incubated for 30 min (F(ab)’2-Goat anti-Human IgG Fc; H10104, Invitrogen). Detection was carried out with a FlexMap 3D analyzer.
In-house ELISA
ELISAs were developed for the detection of IgG/A/M antibodies against the following molecules: PF4, B2-Gp1, and cardiolipin. All ELISAs were developed with clear, high-binding, half-area 96-well plates (734-1624, Corning, VWR). For protein coating, the final protocol was to obtain 1–2 µg/ml protein diluted in PBS containing 0.01% BSA. Fifty µl of protein solution was added to each well, the plate was sealed, and coating was performed overnight at 4 °C. For cardiolipin, the protocol was to obtain 10 µg/ml cardiolipin concentration in 50 µl ethanol (initial purity 99.5%) and the plate was left unsealed at 4 °C overnight for complete evaporation. In the event that complete dryness was not observed on the following day, the plate was left at room temperature for up to 30 min before proceeding with the assay.
The washing buffer was PBS with 0.1% Tween-20, and plate washing was performed in a standard fashion with 130 µl of buffer (5 times). Plates were blocked for 2 h at room temperature using 2% BSA in PBS with 0.01% Tween-20 (55 µl). Samples and positive controls were diluted with PBS (1:2000) in a 2-step process (1:20 then 1:100) before being immediately transferred to wells for incubation (50 µl, 1.5 h at room temperature) with slight agitation achieved on an orbital shaker set to 200 RPM. Secondary antibodies were added at a volume of 50 µl for 1 h (diluted at 1:10000). Different secondary antibodies were used to identify Ig types (IgG/A/M, IgG, and IgM; Invitrogen A18847, A18805, and 31415). Color development was achieved by 5–10 min of 3, 3′, 5, 5′ tetramethylbenzidine (TMB) incubation. The reaction was stopped with 40 µl of 0.2 M H2SO4, and the optical density was recorded at 450 nm (Magellan, TECAN).
Commercial assays
To detect antibodies against PF4-polyanion complexes (PF4C), we used the Lifecodes PF4 Enhanced assay (X-HAT45G, Immucor), which is used for diagnostic purposes in patients with heparin-induced thrombocytopenia and shows excellent sensitivity for VITT35. An additional step to confirm antiphospholipid antibodies (such as B2-Gp1 and cardiolipin) was performed on samples with relatively elevated levels in the bead-based assay or in-house ELISA, by using a commercial screening kit capable of detecting IgG/M autoantibodies (Arigo Biolaboratories, ARG80405, Human Phospholipid Screen IgG/IgM). Protein S levels were measured using an ELISA kit (Novus Biologicals, NBP2-60585, Lot# 101802311), with calculation performed via four-parameter logistic regression.
Confirmatory analyses
For the confirmation of protein S autoantibodies, we employed a separate optimized ELISA on the following samples: the 6 patients with the highest responses in the bead-based assay (confirmation subgroup), 8 randomly-selected patients with myocarditis, and 8 randomly-selected BDs. Protein S was coated at 1.6 µg/ml in PBS, blocking buffer was 3% BSA in PBS with 0.05% Tween-20, samples were added to wells with 1:250 final dilution, and secondary antibody was diluted 1:8000. All other steps of the protocol were the same as described above (in-house ELISA).
Protein S activity was also tested through a manual method, utilizing a functional assay (ACTICLOT Protein S, BioMedica Diagnostics) which outputs a percentage-wise protein S activity value based on sample clotting time. Since the blood samples from patients with AEFIs were collected in heparinized tubes, the assay was not readily applicable to these samples (demonstrated to be working with EDTA and citrated plasma). We utilized protein A and protein G magnetic beads to purify IgG from the heparinized plasma of the 8 patients with the highest immunoreactivity to protein S. These were mixed with pooled EDTA plasma to create assayable samples (Supplementary methods). Time until clot development was kept manually. The reference range for normal protein S activity was 55–160%. The decision to include 8 subjects with available samples in this analysis (instead of only the 6 subjects defined to have positivity) was made to improve data comprehensiveness based on the fact that the bead-based results obtained for these additional patients were very close to the thresholds set for positivity.
Statistics
All data were entered into SPSS (.sav) databases which were imported into Rstudio software using the “haven” and “sjlabelled” packages. Missing data were not imputed and were excluded from analyses. For data visualization, we used the “ggplot2” and “pheatmap” packages for R (version 4.3.0–“Already Tomorrow”; Rstudio release “Ocean Storm”, 2024-01-28)36. To obtain data summaries (descriptives) and perform statistical analyses, we used the SPSS v25.0 (IBM, NY, USA) software. Numerical data were summarized in the form of mean ± SD, while nominal and ordinal data were summarized with absolute (n) and relative frequencies (%). For all categorical variables, we used appropriate Chi-square tests or the Fisher’s Exact test to test for differences in relative distribution between groups. For numerical variables, histograms and Q-Q plots were used to assess normality of distribution, supplemented with the Kolmogorov-Smirnov (Lilliefors correction) test to exclude normality. Comparison of numerical variables between two independent groups was performed with the Mann-Whitney U test, while > 2-group comparisons were performed with one-way ANOVA (parametric) or the Kruskal–Wallis test (non-parametric), with Bonferroni correction used for pairwise analysis. The effect sizes of directional relationships between continuous variables were analyzed by calculating the Pearson correlation coefficient (r).