Cell lines
Expi293F cells are derived from the HEK293F cell line (Life Technologies). Expi293F cells were grown in Expi293 Expression Medium (Life Technologies), cultured at 36.5 °C with 8% CO2 and shaking at 150 rpm. HEK293T/17 is a female human embryonic kidney cell line (ATCC). The HEK-ACE2 adherent cell line was obtained through BEI Resources, NIAID, NIH: Human Embryonic Kidney Cells (HEK293T) Expressing Human Angiotensin-Converting Enzyme 2, HEK293T-hACE2 Cell Line, NR-52511. The VeroE6-TMPRSS2 cell line is an African Green monkey kidney cell line expressing TMPRSS2 (ref. 52). All adherent cells were cultured at 37 °C with 5% CO2 in flasks with DMEM + 10% FBS (Hyclone) + 1% penicillin-streptomycin. Cell lines other than Expi293F were not tested for mycoplasma contamination nor authenticated.
Mice
Four week-old female BALB/c mice (order code 047, BALB/cAnNHsd strain) were obtained from Envigo, and maintained in a specific pathogen-free facility at the University of Washington, Seattle, WA, accredited by the American Association for the Accreditation of Laboratory Animal Care International (AAALAC). Animal procedures were performed under the approvals of the Institutional Animal Care and Use Committee (IACUC) of University of Washington, Seattle, WA. Kymab, a Sanofi Company’s proprietary IntelliSelect™ transgenic mouse platform, known as Darwin, has complete human antibody loci with a non-rearranged human antibody variable and constant germline repertoire. Consequently, the antibodies produced by these mice are fully human.
Plasmid construction
The Wu-1-SARS-CoV-2, B.1.351(β)-SARS-CoV-2, P.1(γ)-SARS-CoV-2, and E484K RBDs (N-RFPN…KKST-C) were genetically fused to the N terminus of the trimeric I53-50A nanoparticle component using a 16-residue glycine and serine linker. The RBD-16GS-I53-50A fusions were cloned into pCMV/R using the Xba1 and AvrII restriction sites using Gibson assembly or by GenScript. All RBD-bearing components contained an N-terminal mu-phosphatase signal peptide and a C-terminal octa-histidine tag. Rpk9 mutants were designed the same way except with mutations at Y365F, F392W, V395I. The Wu-1, B.1.351(β), and P.1(γ)-SARS-CoV-2 S-6P ectodomain trimers were synthesized by GenScript into pCMV/R with an N-terminal BM40 signal peptide and a C-terminal T4 fibritin foldon trimerization domain and octa-histidine tag. The constructs contained the HexaPro mutations (proline substitutions at residues F817P, A892P, A899P, A942P, K986P, and V987P) and 682SGAG685 substitution at the furin cleavage site.
Transient transfection
Proteins were produced using endotoxin-free DNA in Expi293F cells grown in suspension using Expi293F expression medium (Life Technologies) at 33 °C, 70% humidity, 8% CO2 rotating at 150 rpm. The cultures were transfected using PEI-MAX (Polyscience) with cells grown to a density of 3.0 million cells per mL and cultivated for 3 days. Supernatants were clarified by centrifugation (5 min at 4000 rcf), addition of PDADMAC solution to a final concentration of 0.0375% (Sigma Aldrich, #409014), and a second centrifugation (5 min at 4000 rcf).
Microbial plasmid construction, protein expression and purification of I53-50B.4PT1
I53-50B.4PT1 plasmid was synthesized by GenScript in pET29b between the NdeI and XhoI restriction sites with a double-stop codon just before the C-terminal polyhistidine tag. Protein was expressed in Lemo21(DE3) cells (NEB) in LB (10 g Tryptone, 5 g Yeast Extract, 10 g NaCl) grown in a 10 L BioFlo 320 Fermenter (Eppendorf). At inoculation, impeller speed was set to 225 rpm, SPLM set to 5 with O2 supplementation as part of the dissolved-oxygen aeration cascade, and the temperature set to 37 °C. At the onset of a DO spike (OD ~ 12), the culture was fed with a bolus addition of 100 mL of 100% glycerol and induced with 1 mM IPTG. During this time, the culture temperature was reduced to 18 °C, and O2 supplementation was ceased, with expression continuing until an OD ~ 20. The culture was harvested by centrifugation and the pellets were resuspended in PBS, homogenized, and then lysed by microfluidization using a Microfluidics M110P at 18,000 psi using 3 discrete passes. Following sample clarification by centrifugation (24,000 g for 30 min), the supernatant was discarded, and protein was extracted from the inclusion bodies. First, the pellet was washed with PBS supplemented with 0.1% Triton X-100, pH 8.0. After this initial wash and sample clarification by centrifugation, the pellet was washed with PBS supplemented with 1 M NaCl, pH 8.0. Following the second wash, the protein was extracted from the pellet into PBS supplemented with 2 M urea and 0.75% CHAPS (3-((3-Cholamidopropyl)dimethylammonio)-1-propanesulfonate), pH 8.0. Following extraction, the sample was applied to a DEAE Sepharose FF column (Cytiva) on an ӒKTA Avant150 FPLC system (Cytiva). After sample binding, the column was washed with 5 column volumes (CV) of PBS supplemented with 0.1% Triton X-100, pH 8.0, followed by a 5 CV wash with PBS supplemented with 0.75% CHAPs, pH 8.0 in series. The protein was eluted with 3 CV of PBS supplemented with 500 mM NaCl, pH 8.0. After purification, fractions were pooled and concentrated in 10 K molecular weight cutoff (MWCO) centrifugal filters (Millipore), sterile-filtered (0.22 μm), and tested to confirm low endotoxin levels before use in nanoparticle assembly.
Protein purification
Proteins containing His tags were purified from clarified supernatants via a batch bind method where each clarified supernatant was supplemented with 1 M Tris-HCl pH 8.0 to a final concentration of 45 mM and 5 M NaCl to a final concentration of ∼310 mM. Talon cobalt affinity resin (Takara) (for S-6P) or Nickel Sepharose Excel resin (Cytiva) (for VOC-RBD-I53-50A) were added to the treated supernatants and allowed to incubate for 15 min with gentle shaking. Resin was collected using vacuum filtration with a 0.2 μm filter and transferred to a gravity column. The resin was washed with 20 mM Tris pH 8.0, 300 mM NaCl, and the protein was eluted with 3 CV of 20 mM Tris pH 8.0, 300 mM NaCl, 300 mM imidazole. The batch bind process was then repeated and the first and second elutions combined. SDS-PAGE was used to assess purity. VOC-RBD-I53-50A fusion protein IMAC elutions were concentrated to >1 mg/mL and subjected to three rounds of dialysis into 50 mM Tris pH 7.4, 185 mM NaCl, 100 mM L-arginine, 4.5% glycerol, and 0.75% w/v CHAPS (MAGiC Sauce, MS or Mixture of Arginine, Glycerol, in CHAPS) in a hydrated 10 K MWCO dialysis cassette (Thermo Scientific). VOC-HexaPro elutions were concentrated to >0.5 mg/mL and subjected to three rounds of dialysis into 50 mM Tris pH 8, 185 mM NaCl, 0.25% w/v L-histidine, 5% v/v glycerol in a hydrated 10 K MWCO dialysis cassette (Thermo Scientific). Clarified supernatants of cells expressing monoclonal antibodies and human ACE2-Fc were purified using a MabSelect PrismA 2.6 × 5 cm column (Cytiva) on an ӒKTA Avant150 FPLC (Cytiva). Bound antibodies were washed with five CV of 20 mM NaPO4, 150 mM NaCl pH 7.2, then five CV of 20 mM NaPO4, 1 M NaCl, pH 7.4 and eluted with three CV of 100 mM glycine at pH 3.0. The eluate was neutralized with 2 M Trizma base to 50 mM final concentration. SDS-PAGE was used to assess purity.
In vitro nanoparticle assembly and purification
Total protein concentration of purified individual nanoparticle components was determined by measuring absorbance at 280 nm using a UV/vis spectrophotometer (Agilent Cary 8454) and calculated extinction coefficients. The assembly steps were performed at room temperature with addition in the following order: VOC-RBD-I53-50A trimeric fusion protein, followed by additional buffer (50 mM Tris pH 7.4, 185 mM NaCl, 100 mM L-arginine, 4.5% glycerol, and 0.75% w/v CHAPS) as needed to achieve desired final concentration, and finally I53-50B.4PT1 pentameric component (in 50 mM Tris pH 8, 500 mM NaCl, 0.75% w/v CHAPS), with a molar ratio of VOC-RBD-I53-50A:I53-50B.4PT1 of 1.1:1. All VOC-RBD-NP in vitro assemblies were incubated briefly at room temperature before subsequent purification by SEC in order to remove residual unassembled VOC-RBD-I53-50A component. A Superose 6 Increase 10/300 GL column was used for nanoparticle purification. Assembled nanoparticles were purified in 50 mM Tris pH 7.4, 185 mM NaCl, 100 mM Arginine, 4.5% v/v glycerol, and 0.75% w/v CHAPS, and elute at ∼11 mL on the Superose 6 column. Assembled nanoparticles were sterile-filtered (0.22 μm) immediately prior to column application and following pooling of fractions.
UV/vis
UV/vis absorbance was measured using an Agilent Technologies Cary 8454. Samples were applied to a 10 mm, 50 μL quartz cell (Starna Cells, Inc.) and absorbance was measured from 180 to 1000 nm. Net absorbance at 280 nm, obtained from measurement and single reference wavelength baseline subtraction, was used with calculated extinction coefficients and molecular weights to obtain protein concentration. The ratio of absorbance at 320/280 nm was used to determine relative aggregation levels in real-time stability study samples. Samples were diluted with respective purification/instrument blanking buffers to obtain an absorbance between 0.1 and 1.0. All data produced from the UV/vis instrument was processed in the 845x UV/visible System software.
Endotoxin measurements
Endotoxin levels in protein samples were measured using the EndoSafe Nexgen-MCS System (Charles River). Samples were diluted 1:100 in Endotoxin-free LAL reagent water, and applied into wells of an EndoSafe LAL reagent cartridge. Charles River EndoScan-V software was used to analyze endotoxin content, automatically back-calculating for the dilution factor. Endotoxin values were reported as EU/mL which were then converted to EU/mg based on UV/vis measurements. Our threshold for samples suitable for immunization was <50 EU/mg.
Dynamic light scattering
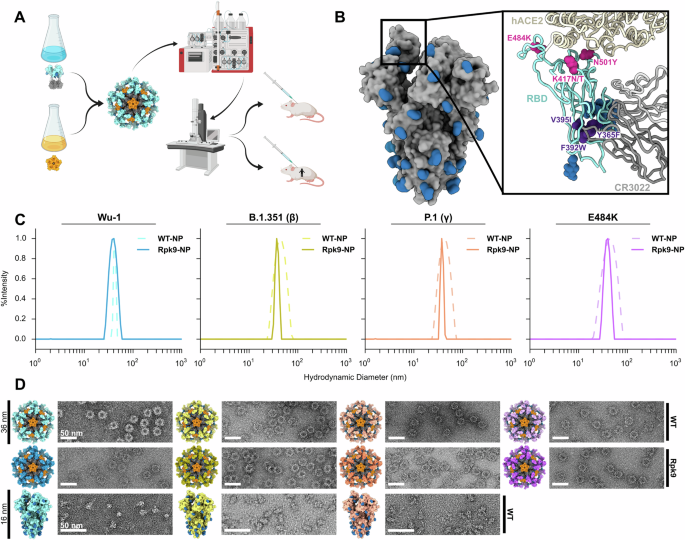
Dynamic light scattering (DLS) was used to measure hydrodynamic diameter (Dh) and % Polydispersity (%Pd) of RBD-NPs, mRBD-NPs, and cRBD-NPs on an UNcle Nano-DSF (UNchained Laboratories). Sample was applied to a 8.8 µL quartz capillary cassette (UNi, UNchained Laboratories) and measured with 10 acquisitions of 5 s each, using auto-attenuation of the laser. Increased viscosity due to 4.5% v/v glycerol in the RBD nanoparticle buffer was accounted for by the UNcle Client software in Dh measurements.
Negative stain electron microscopy
VOC-RBD-NP and VOC-HexaPro vaccines were first diluted to 75 µg/mL or 30 µg/mL in 50 mM Tris pH 7.4, 185 mM NaCl, 100 mM L-arginine, 4.5% v/v glycerol, 0.75% w/v CHAPS or 50 mM Tris pH 8, 185 mM NaCl, 0.25% w/v L-histidine, 5% v/v glycerol prior to application of 3 µL of sample onto freshly glow-discharged 300-mesh copper grids. Sample was incubated on the grid for 1 min before 6 µL of 0.75% w/v uranyl formate stain was applied to the grid. Stain was blotted off with filter paper, then the grids were dipped into another 6 µL of stain and then repeated once more. Finally, the stain was blotted away and the grids were allowed to dry for 1 min. Prepared grids were imaged in a Talos model L120C electron microscope at 57,000× or 92,000×.
hACE2-Fc, CR3022, and LYCoV555 IgG digestion
hACE2-Fc was digested with thrombin protease (Sigma Aldrich) in the presence of 2.5 mM CaCl2 at a 1:300 w/w thrombin:protein ratio. The reaction was incubated at ambient temperature for 16–18 h with gentle rocking. Following incubation, the reaction mixture was concentrated using Ultracel 10 K MWCO centrifugal filters (Millipore Amicon Ultra) and sterile filtered (0.22 μM). Cleaved hACE2 monomer was separated from uncleaved hACE2-Fc and the cleaved Fc regions using Protein A purification (see Protein purification above) on a HiScreen MabSelect SuRe column (Cytiva) using an ӒKTA avant 25 FPLC (Cytiva). Cleaved hACE2 monomer was collected in the flowthrough, sterile-filtered (0.22 μm), and quantified by UV/vis.
LysC (New England BioLabs) was diluted to 10 ng/μL in 10 mM Tris pH 8 and added to CR3022 or LYCoV555 IgG at 1:2000 w/w LysC:IgG and subsequently incubated for 18 h at 37 °C with orbital shaking at 230 rpm. The cleavage reaction was concentrated using Ultracel 10 K MWCO centrifugal filters (Millipore Amicon Ultra) and sterile filtered (0.22 μM). Cleaved CR3022 or LYCoV555 mAb was separated from uncleaved CR3022 or LYCoV555 IgG and the Fc portion of cleaved IgG, using Protein A purification as described above. Cleaved CR3022 or LYCoV555 was collected in the flowthrough, sterile-filtered (0.22 μm), and quantified by UV/vis.
Bio-layer interferometry (affinity determination)
Affinity assays were performed and analyzed using BLI on an Octet Red 96 System (Pall Forté Bio/Sartorius) at ambient temperature with shaking at 1000 rpm. VOC-RBD-I53-50A trimeric components were diluted to 40 μg/mL in Kinetics buffer (1× HEPES-EP+ (Pall Forté Bio), 0.05% nonfat milk, and 0.02% sodium azide). Monomeric hACE2, CR3022, and LYCoV555 Fab were diluted to 750 or 300 nM in Kinetics buffer and serially diluted three-fold for a final concentration of 3.1 or 1.23 nM. Reagents were applied to a black 96-well Greiner Bio-one microplate at 200 μL per well as described below. VOC-RBD-I53-50A components were immobilized onto Anti-Penta-HIS (HIS1K) biosensors per manufacturer instructions (Forté Bio) except using the following sensor incubation times. HIS1K biosensors were hydrated in water for 10 min, and were then equilibrated in Kinetics buffer for 60 s. The HIS1K tips were loaded with diluted trimeric VOC-RBD-I53-50A component or monomeric RBD for 150 s and washed with Kinetics buffer for 60 s. The association step was performed by dipping the HIS1K biosensors with immobilized immunogen into diluted hACE2 monomer, CR3022, or LYCoV555 Fab for 200 s, then dissociation was measured by inserting the biosensors back into Kinetics buffer for 200 s. The data were baseline subtracted and the plots fitted using the Pall FortéBio/Sartorius analysis software (version 12.0). Plots in Fig. 2 show the association and dissociation steps.
ELISA (antigenicity)
For anti-HexaPro ELISA, 50 μL of 2 μg/mL HexaPro S was plated onto 384-well Nunc Maxisorp (ThermoFisher) plates in PBS and sealed overnight at RT. The next day, the plates were slapped dry and blocked with Casein (ThermoFisher) for 1 h at 37 °C. Plates were washed 4× in Tris-Buffered Saline Tween (TBST) using a plate washer (BioTek) and 1:5 serial dilutions of mAb were made in 40 μL TBST and incubated at 37 °C for 1 h. Plates were washed 4× with TBST on the BioTek plate washer, then anti-human (Invitrogen) horseradish peroxidase-conjugated antibodies were diluted 1:5,000 and 40 μL added to each well and incubated at 37 °C for 1 h. Plates were washed 4× in TBST on the BioTek plate washer and 40 μL of room temperature TMB (SeraCare) was added to every well for ∼5 min at room temperature. The reaction was quenched with the addition of 40 μL of 1 N HCl. Plates were immediately read at 450 nm on a BioTek plate reader and data plotted and fit in Prism (GraphPad) using nonlinear regression sigmoidal, 4PL, X is log(concentration) to determine EC50 values from curve fits. ELISA experiments were performed three separate times and representative experiments are shown.
Hydrogen/Deuterium-exchange mass spectrometry
3 μg of VOC-RBD-I53-50A and VOC-Rpk9-RBD-I53-50A mutants were H/D exchanged in the deuteration buffer (pH* 7.5, 85% D2O, Cambridge Isotope Laboratories, Inc.) at 23 °C for 3, 60, 1800, and 72000 s, respectively. H/D exchanged samples were immediately mixed with an equal volume of ice-cold quench buffer (8 M urea, 200 mM tris(2-chloroethyl) phosphate (TCEP), 0.2% formic acid (FA)) to a final pH 2.5 and flash frozen in liquid nitrogen. Samples were analyzed by LC-MS on a Waters Synapt G2-Si mass spectrometer using a custom-built cooling box to maintain sample digestion and injection at 0 °C. Quenched samples were loaded over an immobilized pepsin column (2.1 × 50 mm) with a 200 μL/min flow of loading buffer (0.1% trifluoroacetic acid (TFA), 2% acetonitrile (ACN)). Peptides were trapped on a Waters CSH C18 trap cartridge (2.1 × 5 mm) and resolved over a Waters CSH C18 1 × 100 mm 1.7 μm column with a linear gradient from 3% to 40% B over 18 min (A: 2% ACN, 0.1% FA, 0.025% TFA; B: 100% ACN, 0.1% FA, flow rate of 40 μL/min). A series of washes steps were performed between all samples to minimize carryover53. The total deuterated control was made by collecting a pepsin digest elution, drying by speed-vacuum, and incubating in deuteration buffer for 1 h at 85 °C. Peptides were identified and updated based on our previously examined lists20,54 and validated using DriftScope™ (Waters). Deuterium uptake analysis was performed with HDExaminer (Sierra Analytics) and HX-Express v255,56. Peaks were identified from the peptide spectra with binomial fitting applied. The deuterium uptake level was normalized relative to total deuterated control.
Nanoscale differential scanning fluorimetry (Component and NP)
nanoDSF was used to measure the relative melting temperature (Tm) and aggregation (Tagg) temperatures of the individual VOC-RBD-I5350As and VOC-RBD-NPs, respectively, on an UNcle Nano-DSF (UNchained Laboratories). VOC-RBD-I5350As and VOC-RBD-NPs were diluted to 0.5 mg/mL in MAGiC Sauce (50 mM Tris pH 7.4, 185 mM NaCl, 100 mM L-arginine, 4.5% glycerol, and 0.75% w/v CHAPS) and 8.8 µL were applied to a 8.8 µL quartz capillary cassette (UNi, UNchained Laboratories). The barycentric mean (BCM) and static light scattering at 488 nm (SLS 488 nm) were taken from 20 °C to 95 °C. Tm and Tagg were approximated by analyzing when the derivative or second derivative of the line fit for BCM or SLS 488 nm, respectively, was approximately equal to zero.
Bio-layer interferometry
Binding of hACE2-Fc to monovalent VOC-RBD-NP, mVOC-RBD-NP, and cVOD-RBD-NP was analyzed for antigenicity experiments and real-time stability studies using an Octet Red 96 System (Pall FortéBio/Sartorius) at ambient temperature with shaking at 1000 rpm. Protein samples were diluted to 100 nM in Kinetics buffer (1 × HEPES-EP+ (Pall Forté Bio), 0.05% nonfat milk, and 0.02% sodium azide). Buffer, receptor, and analyte were then applied to a black 96-well Greiner Bio-one microplate at 200 μL per well. Protein A biosensors (FortéBio/Sartorius) were first hydrated for 10 min in Kinetics buffer, then dipped into hACE2-Fc diluted to 10 μg/mL in Kinetics buffer in the immobilization step. After 150 s, the tips were transferred to Kinetics buffer for 60 s to reach a baseline. The association step was performed by dipping the loaded biosensors into the immunogens for 200 s, and subsequent dissociation was performed by dipping the biosensors back into Kinetics buffer for an additional 200 s. The data were baseline subtracted prior for plotting using the FortéBio analysis software (version 12.0).
BALB/c mice immunizations
At 8 weeks of age, 5 female BALB/c mice per dosing group were vaccinated with a prime immunization, and 3 weeks later mice were boosted with a second vaccination (IACUC protocol 4470.01). Prior to inoculation, immunogen suspensions were gently mixed 1:1 vol/vol with AddaVax adjuvant (Invivogen, San Diego, CA) to reach a final concentration of 0.01 mg/mL antigen and 0.13125% w/v CHAPS. Mice were injected intramuscularly into the gastrocnemius muscle of each hind leg using a 27-gauge needle (BD, San Diego, CA) with 50 μL per injection site (100 μL total) of immunogen under isoflurane anesthesia. Specifically, mice underwent induction of anesthesia in an induction chamber using 3–5% isoflurane with an O2 flow rate of 1 L/min. After assessing depth of anesthesia via toe pinch, mice were transferred from induction chambers into nosecones with the same isoflurane and oxygen parameters for immunization. Euthermia was maintained throughout the procedure using circulating warm water pads. Following immunization, mice recovered from anesthesia in separate recovery chambers. To obtain sera all mice were bled 2 weeks after prime and boost immunizations. Blood was collected via submental venous puncture and rested in 1.5 mL plastic Eppendorf tubes at room temperature for 30 min to allow for coagulation. Serum was separated from red blood cells via centrifugation at 2000 g for 10 min. Complement factors in isolated serum were heat-inactivated via incubation at 56 °C for 60 min. Serum was stored at 4 °C or −80 °C until use. After the post-boost blood collection, mice were euthanized in CO2 chambers with secondary confirmation via cervical dislocation. BALB/c immunogenicity studies were repeated twice.
Darwin mice immunizations
Kymab Darwin mice, equivalent to the Kymouse31 in the IgH locus except the immunoglobulin constant regions are also fully human, and with a fully human IgL kappa loci, were used in this study (a mix of males and females, 10 weeks of age). Five mice per dosing group were vaccinated with a prime immunization and 3 weeks later boosted with a second vaccination. Prior to inoculation, immunogen suspensions were gently mixed 1:1 vol/vol with AddaVax adjuvant (Invivogen) to reach a final concentration of 0.025 mg/mL antigen. Mice were injected intramuscularly into the tibialis muscle of each hind leg using a 30-gauge needle (BD) with 20 μL per injection site (40 μL total) of immunogen under isoflurane anesthesia. A final boost was administered intravenously (50 μL) with no adjuvant at week 7 in order to recruit circulating B cells to the spleen. Mice were sacrificed 5 days later under UK Home Office Schedule 1 (rising concentration of CO2) and spleen, lymph nodes, and bone marrow cryopreserved. Whole blood (0.1 mL) was collected 7.5 weeks after the first immunization. Serum was separated from hematocrit via centrifugation at 2000 g for 10 min. Serum was stored at −20 °C. All mice were maintained and all procedures carried out under United Kingdom Home Office License 70/8718 and with the approval of the Wellcome Trust Sanger Institute Animal Welfare and Ethical Review Body.
Pseudovirus production
SARS-CoV-2 Wu-1 G614 S, B.1.351 S, P1, B.1.617.2 S, and B.1.1.259.BA.1 S pseudotyped VSV viruses were prepared as described previously57,58,59. Briefly, 293 T cells in DMEM supplemented with 10% FBS, 1% PenStrep seeded in polylysine coated 10-cm dishes were transfected with the plasmid encoding for the corresponding S glycoprotein using lipofectamine 2000 (Life Technologies) following manufacturer’s indications. One day post-transfection, cells were infected with VSV(G∗ΔG-luciferase) and after 2 h were washed five times with warm DMEM before adding medium supplemented with anti-VSV-G antibody (I1- mouse hybridoma supernatant, CRL- 2700, ATCC). Virus pseudotypes were harvested 18–24 h post-inoculation, clarified by centrifugation at 2500 g for 10 min, filtered through a 0.45 μm cutoff membrane, concentrated 10 times with a 30 kDa MWCO Sartorius membrane, aliquoted, and stored at −80 °C prior to use at a 1:25 dilution in DMEM.
Pseudovirus neutralization
HEK293-hACE2 cells60 were cultured in DMEM with 10% FBS (Hyclone) and 1% PenStrep with 8% CO2 in a 37 °C incubator (ThermoFisher). One day prior to infection, 40 μL of poly-lysine (Sigma) was placed into white 96-well plates and incubated with rotation for 5 min. Poly-lysine was removed, plates were dried for 5 min then washed 1× with water prior to plating with ~40,000 cells. The following day, cells were checked to be at 80% confluence. In an empty half-area 96-well plate, a 1:3 serial dilution of sera was made in DMEM and diluted pseudovirus was then added to the serial dilution and incubated at room temperature for 30–60 min. After incubation, the sera-virus mixture was added to the cells and incubated at 37 °C for 2 hours. Following the 2 h incubation, 40 μL of 20% FBS-2% PenStrep DMEM was added for overnight incubation. After 16–20 h 40 μL/well of One-Glo-EX substrate (Promega) was added to the cells and incubated in the dark for 5–10 min prior reading on a BioTek plate reader. Measurements were done in at least duplicate with distinct pseudovirus batches and backbones. Relative luciferase units were plotted and normalized in Prism (GraphPad). Nonlinear regression of log(inhibitor) versus normalized response was used to determine IC50 values from curve fits.
Quantification and statistical analysis
Statistical details of experiments can be found in the figure legends. For mouse experiments, 5 BALB/c animals were used and experiments were completed in at least duplicate. Geometric mean titers were calculated. Kruskal–Wallis tests were performed to compare two groups to determine whether they were statistically different. Significance is indicated with stars: ∗p < 0.05; ∗∗p < 0.01 and non-significant differences are not shown.