Mice
Six-week-old female BALB/c mice were purchased from SLC (Hamamatsu, Shizuoka, Japan). They were housed under a 12-hour light/dark cycle (lights on at 8:00 AM, lights off at 8:00 PM) with ad libitum access to food and water. All animal procedures were approved by the Animal Care and Use Committee of the Research Institute for Microbial Diseases, Osaka University, Japan (protocol numbers: BIKEN-AP-R01-15-2 and BIKEN-AP-R02-14-5) and were performed according to Osaka University’s Institutional Guidelines for the Ethical Treatment of Animals.
Production and purification of recombinant G protein and IL-13Rα2-Fc protein
For G protein expression, the G protein sequence was derived from RSV (strain A2) (GenBank accession number: AAB59857.1). The plasmid vector was encoded using the human codon-optimized sequence of the human Igκ signal peptide (METDTLLLWVLLLWVPGSTGD), hexahistidine tag (His-tag), linker (GAGGG), and G protein ectodomain (amino acids 67–298). For expression of IL-13Rα2-Fc protein, the plasmid vector was encoded with the human codon-optimized sequence of human Igκ signal peptide, the extracellular domain of mouse IL-13Rα2 (GenBank accession number: AAC33240.1) (amino acids 22–334), G4S linker (GGGGS), His-tag, G4S linker, and the hinge-CH2-CH3 regions of human IgG1 (GenBank accession number: AAC82527.1) (amino acids 100–330). Recombinant G and IL-13Rα2-Fc proteins were expressed in Expi293F cells (Thermo Fisher Scientific, Hampton, NH, USA) and purified by immobilized metal affinity chromatography and size exclusion chromatography, as previously reported12.
RSV progression and enrichment
RSV (strain A2) was produced using HEp-2 cells as previously reported12. Briefly, for RSV progression, one plaque forming unit (PFU) of RSV per 102 HEp-2 cells was added to 70–80% confluent HEp-2 monolayers and incubated at 37 °C with 5% CO2 for 7 h with shaking every hour. The culture medium was then replaced with fresh medium. After incubation at 37 °C with 5% CO2 for 5 days, the cells were freeze-thawed to collect the virus, and the supernatant was collected by centrifugation twice at 700 × g for 5 min at 4 °C. For RSV enrichment, the supernatant was transferred to an ultracentrifuge tube, to which 1 mL of PBS containing 20% sucrose was added, and centrifuged at 71,000 × g for 3.5 h at 4 °C. The precipitated virus was resuspended in PBS, aliquoted, and stored at –80 °C until use. Virus titers were measured using a plaque assay as previously reported12. Briefly, the virus diluent was added to 90% confluent HEp-2 monolayers and incubated at 37 °C with 5% CO2 for 2 h. Following the incubation, the culture medium was replaced with fresh medium containing 0.6% carboxymethylcellulose. Subsequently, the cells and viruses were fixed with methanol after incubation at 37 °C with 5% CO2 for 3 days. The fixed viruses were then treated with goat anti-RSV polyclonal antibody (catalog number: AB1128, dilution 1/500; Merck Millipore, Darmstadt, Germany) and horseradish peroxidase-conjugated donkey anti-goat IgG polyclonal antibody (catalog number: A15999, dilution 1/150; Thermo Fisher Scientific). The plaques were visualized using 4-chloro-1-napthol (Tokyo Chemical Industry Co., Ltd., Tokyo, Japan) and subsequently counted. All animal experiments were reviewed and approved by the Institutional Review Board of the Research Institute for Microbial Diseases, Osaka University (protocol number: BIKEN-00224-002).
Vaccination and RSV challenge
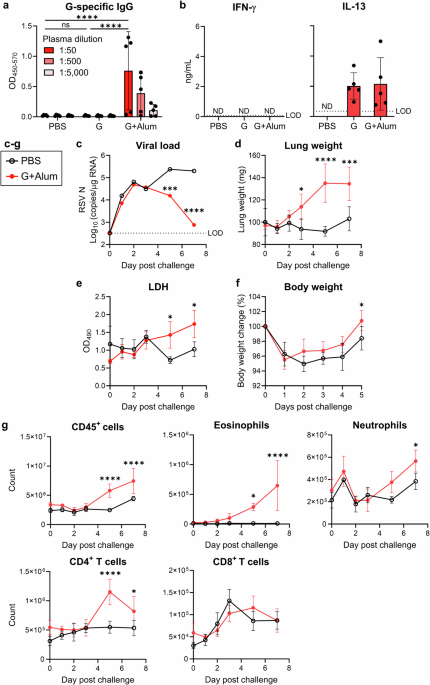
Mice were vaccinated subcutaneously with 1 μg of G protein, either alone or with 50 μg of Alhydrogel adjuvant 2% (Alum; InvivoGen, San Diego, CA, USA), in 50 μL of PBS at the base of the tail on days 0 and 21. On day 28, plasma samples were collected from the cheek vein to assess G-specific IgG levels. Following euthanasia via cervical dislocation, spleens were harvested for the T-cell restimulation assay, as previously described12. On day 31, mice were intranasally challenged with 1.0 × 105 PFU of RSV in 30 μL of PBS (15 μL to each nostril) under anesthesia. After RSV challenge, the mice were anesthetized and euthanized. Lungs and BALF samples were collected for use in subsequent assays. In the experiments shown in Figs. 1d, 2a, 2d, 3a, 3d, and 5d, the weight of the right lung was measured. In the experiments shown in Figs. 5b, 5c, and 6c, the weight of both lungs was measured.
Detection of antibodies and cytokines
Plasma levels of G-specific IgG were detected using an indirect enzyme-linked immunosorbent assay (ELISA), as previously reported12. Briefly, 96-well half-area flat-bottom plates (Corning, NY, USA) were coated with 1 μg/mL G protein in carbonate buffer. Diluted plasma was added to G protein-coated wells. Antibodies bound to G protein were incubated with the antibodies listed in Supplementary Table 1. The coloration reaction was performed using tetramethylbenzidine, followed by stopping with 2 N H2SO4. The difference in optical density (OD) between 450 and 570 nm (OD450-570) was detected using a microplate reader (Power Wave HT; BioTek, Winooski, VT, USA). Cytokine levels were detected using a sandwich ELISA with the reagents listed in Supplementary Table 1. IL-13 levels were determined according to the manufacturer’s instructions using an IL-13 Mouse Uncoated ELISA Kit (Thermo Fisher Scientific). OD450-570 was measured using a microplate reader.
Detection of mRNA expressions
The right lungs were collected in 1 mL TRIzol Reagent (Thermo Fisher Scientific) and homogenized with three 4 mm stainless steel beads (TAITEC, Saitama, Japan) using a bead beater-type homogenizer (Beads Crusher μT-12; TAITEC) for 60 s at 3200 rpm. RNA was purified using TRIzol Reagent according to the manufacturer’s instructions. Reverse transcription was performed using ReverTra Ace qPCR RT Master Mix (Toyobo, Osaka, Japan) to synthesize cDNA. Real-time RT-PCR was performed by amplifying the target mRNA and Gapdh mRNA as a reference gene using a Light Cycler 480-II (Roche Diagnostics, Tokyo, Japan) and LightCycler 480 SYBR Green I Master (Roche Diagnostics), with the primers listed in Supplementary Table 2. The absolute levels of RSV were calculated by amplifying the plasmid encoding the RSV nucleoprotein (RSV N) gene and generating a standard curve. Relative mRNA expression levels were calculated by dividing the target mRNA expression levels by Gapdh mRNA expression levels, with the mean value of the control group expressed as 1.
LDH assay
BALF (1 mL) was centrifuged at 600 × g for 5 min, and LDH levels in the supernatant were measured using the Cytotoxicity LDH Assay Kit-WST (Dojindo, Kumamoto, Japan) according to the manufacturer’s instructions. OD at 490 nm (OD490) using a microplate reader.
Analysis of infiltrating cells into the lungs
Cells infiltrating the lungs were analyzed as previously reported12. Briefly, the left lungs were sheared and digested using collagenase IV and DNase I. Then, the lungs were dissociated and hemolyzed. For surface antigen staining, lung cells were labeled with the Fixable Viability Dye eFluor 780 (catalog number: 65-0865-18, dilution 1/1000; Thermo Fisher Scientific), and the antibodies are listed in Supplementary Table 3. Flow cytometric analysis was performed using an Attune NxT Flow Cytometer (Thermo Fisher Scientific), and data were analyzed using FlowJo software version 10.10 (FlowJo LLC, Ashland, Oregon, USA).
Depletion of eosinophils, neutrophils, and CD4+ T cells
Mice were vaccinated with the G protein, with or without alum, on days 0 and 21. On day 31, mice were challenged with RSV. For the depletion of CD4+ T cells, mice were injected intraperitoneally on day 30 with 100 μg of rat anti-mouse CD4 antibody (clone: GK1.5, catalog number: BE0003-1; Bio X Cell, West Lebanon, NH, USA) or rat IgG2b isotype control (clone: LTF-2, catalog number: BE0090; Bio X Cell) in 200 μL PBS. For the depletion of eosinophils, mice were injected intraperitoneally on day 30 and 33 with 100 μg of rat anti-mouse CCR3 antibody (clone: 6S2-19-4, catalog number: BE0316; Bio X Cell) or rat IgG2b isotype control in 200 μL PBS. For the depletion of neutrophils, mice were injected intraperitoneally on day 30, 32, and 34 with 50 μg of rat anti-mouse Ly6G antibody (clone: 1A8, catalog number: 127649; BioLegend) or rat IgG2a isotype control (clone: RTK2758, catalog number: 400565; BioLegend) in 200 μL PBS. The right lungs were weighed and collected to measure the viral load. The left lung was collected for analysis of infiltrating cells.
Histopathology analysis
The excised lungs were fixed by injecting 10% phosphate-buffered formalin into the trachea until the lungs were inflated. The left lung of the fixed lung was then embedded in paraffin, sectioned, and stained with H&E or PAS. The histopathological scoring method utilized an ordinal scale with “1” indicating normal or naive status and “4” indicating extensive or severe status, as previously reported58. Specific scoring criteria were as follows: PVA: 1, normal, within naive parameters; 2, focal to uncommon numbers of solitary cells with uncommon aggregates; 3, multifocal moderate aggregates; 4, moderate to high cellularity and multifocal, large cellular aggregates that may be expansive into adjacent tissues. Specific scoring criteria were as follows: mucin: 1, no mucin; 2, goblet cell hyperplasia with none to rare luminal mucin; 3, goblet cell hyperplasia with luminal mucin accumulation in airways; 4, severe mucin alterations, with some airways completely obstructed by mucin.
RNA-seq
Total RNA was extracted from the right lung using a QuickGene-AutoS RNA cultured cell kit (Kurabo, Osaka, Japan) according to the manufacturer’s instructions. RNA libraries were prepared using a TruSeq Stranded mRNA Library Prep Kit (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. Sequencing was performed on NovaSeq 6000 platform in a 101-base single-end mode. RTA v3.4.4 software (Illumina) was used for base calling. Generated reads were mapped to the mouse (mm10) reference genome using TopHat v2.1.1 in combination with Bowtie2 ver. 2.2.8 and SAMtools ver. 0.1.18. Fragments per kilobase of exon per million mapped fragments (FPKMs) were calculated using Cuffdiff 2.2.1 with parameter-max-bundle-frags 50,000,000. Principal component analysis, heatmap clustering, and volcano plot analysis were performed using iDEP 2.01 (http://bioinformatics.sdstate.edu/idep/). Gene set enrichment analysis was performed using GSEA v4.3.3. Dot plots of gene set enrichment analysis were generated using the R function ggplots2. The RNA-seq data concerning this study have been deposited in the Gene Expression Omnibus (GEO) under the accession number GSE272499.
Analysis of T cells infiltrating the lung
The cells isolated from the left lungs (1–3 × 106 cells/well) were cultured in Roswell Park Memorial Institute 1640 medium with 10% fatal bovine serum (FBS), 1% penicillin, 1% streptomycin, and 50 µM 2-mercaptoethanol. The cells were stimulated with 100 ng/mL phorbol 12-myristate 13-acetate and 2 μg/mL ionomycin in presence of protein transport inhibitor cocktail (Thermo Fisher Scientific) for 4 h at 37 °C with 5% CO2 in 96-well U-bottom plates. For surface antigen staining, cells were incubated with Fixable Viability Dye eFluor 780, and the antibodies are listed in Supplementary Table 3. Subsequently, for intracellular cytokine staining, the cells were permeabilized and incubated with the antibodies (Supplementary Table 3) using the BD Cytofix/Cytoperm Fixation/Permeabilization Solution Kit (BD Biosciences, Sparks, MO, USA) according to the manufacturer’s instructions. Flow cytometric analysis was performed using an Attune NxT Flow Cytometer, and data were analyzed using FlowJo software version 10.10.
Intranasal administration of Th2 cytokines
Mice infected or uninfected intranasally with 1.0 × 105 PFU of RSV on the previous day were intranasally administered 1 μg of mouse IL-4 (BioLegend), mouse IL-5 (BioLegend), or mouse IL-13 (BioLegend) in 30 μL PBS (15 μL for each nostril) under anesthesia daily for 4 days. The mice were anesthetized and euthanized on the day after the last dose. The whole lungs were weighed, and the right lungs were collected to measure the mRNA expression levels.
IL-13 neutralization
Mice were vaccinated with G protein on days 0 and 21. On day 31, the mice were challenged intranasally with 1.0 × 105 PFU of RSV in 30 μL of PBS (15 μL to each nostril) under anesthesia. On days 32, 34, and 35, the mice were injected intraperitoneally with 100 μg of IL-13Rα2-Fc in 200 μL PBS. On day 36, mice were anesthetized and euthanized. The right lungs were weighed and collected to measure the viral load. The left lung was collected for analysis of infiltrating cells.
Intranasal administration of NAC
Mice vaccinated with the G protein alone were intranasally administered 1 mg of NAC dissolved in 30 μL PBS (15 μL in each nostril) under anesthesia on day 5 after the RSV challenge. On the day after NAC administration, the mice were anesthetized and euthanized, and their whole lungs were weighed.
Statistical analysis
Statistical analyses were performed using the GraphPad Prism 10 software (GraphPad Software, San Diego, CA, USA). Data are expressed as the means ± standard deviation (SD) or as medians. Significant differences were determined using unpaired Student’s t test, one-way ANOVA with Dunnett’s test or Tukey’s test, or two-way ANOVA with Sidak’s test or Tukey’s test. Statistical significance was defined as P < 0.05.